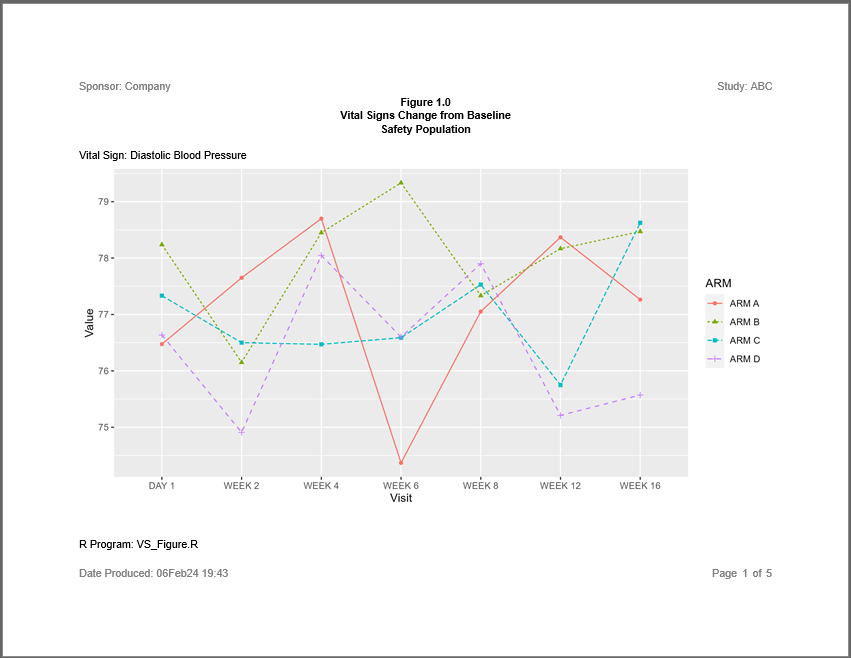
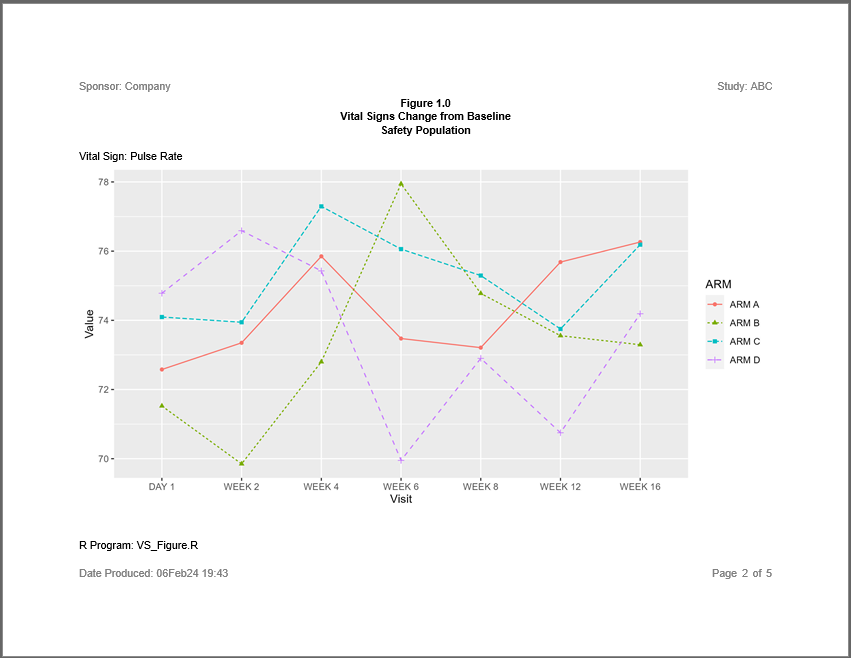
The sassy system gives you capabilities that few other R packages can match. The system not only support reports with by-groups. You can even apply a by-group to a figure.
Program
Note the following about this example:
- The plot is created as a single plot with no by-groups
- The plot is added to the report with the
add_content()function, just like the figures in the previous example. - The
page_by()function on thecreate_plot()statement generates the paging for both the report and plot.
library(sassy)
library(ggplot2)
options("logr.autolog" = TRUE,
"logr.notes" = FALSE)
# Get path to temp directory
tmp <- tempdir()
# Get path to sample data
pkg <- system.file("extdata", package = "sassy")
# Open log
lgpth <- log_open(file.path(tmp, "example6.log"))
# Prepare Data ------------------------------------------------------------
sep("Prepare Data")
put("Load data files")
libname(dat, pkg, "csv", filter = c("DM", "VS"))
put("Prepare factor levels")
visit_levels <- c("SCREENING", "DAY 1", "WEEK 2", "WEEK 4", "WEEK 6",
"WEEK 8", "WEEK 12", "WEEK 16") |> put()
arm_levels <- c("ARM A", "ARM B") |> put()
test_codes <- c("SYSBP", "DIABP", "PULSE", "RESP") |> put()
put("Prepare data for analysis")
datastep(dat$DM, merge = dat$VS, merge_by = v(STUDYID, USUBJID),
keep = v(STUDYID, USUBJID, ARM, VISIT, VSTESTCD, VSORRES),
merge_in = v(inDM, inVS),
where = expression(VISIT != "END OF STUDY EARLY TERMINATION" &
ARM %in% c("ARM A", "ARM B") &
VSTESTCD %in% test_codes
),
{
# Assign factors to VISIT and ARM
VISIT <- factor(VISIT, levels = visit_levels)
ARM <- factor(ARM, levels = arm_levels)
VSTESTCD <- factor(VSTESTCD, levels = test_codes)
if (!inDM & inVS) {
delete()
}
}) -> vitals
# Create Plot -------------------------------------------------------------
sep("Create Plot")
put("Assign Colors")
arm_cols <- c("ARM B" = "#1f77b4", "ARM A" = "#2f2f2f")
put("Define Boxplot")
p_box <- ggplot2::ggplot(
vitals,
ggplot2::aes(x = VISIT, y = VSORRES, fill = ARM, colour = ARM)
) +
ggplot2::geom_boxplot(
position = ggplot2::position_dodge(width = 0.75),
width = 0.6,
outlier.size = 0.7,
alpha = 0.9
) +
ggplot2::scale_fill_manual(values = arm_cols) +
ggplot2::scale_colour_manual(values = arm_cols) +
ggplot2::labs(
x = NULL,
y = "Lab Value",
fill = NULL,
colour = NULL
) +
ggplot2::theme_bw(base_size = 10) +
ggplot2::theme(
legend.position = "bottom",
panel.grid.major.x = ggplot2::element_blank(),
plot.title = ggplot2::element_text(face = "bold", hjust = 0),
plot.caption = ggplot2::element_text(hjust = 0)
)
put("Create format for lab codes")
lbfmt <- value(condition(x == "SYSBP", "Systolic Blood Pressure (mmHg)"),
condition(x == "DIABP", "Diastolic Blood Pressure (mmHg)"),
condition(x == "PULSE", "Pulse (bpm)"),
condition(x == "RESP", "Respirations (bpm)"))
# Report ------------------------------------------------------------------
sep("Report")
put("Create plot object definition")
plt <- create_plot(p_box, height = 4, width = 7, borders = "outside") |>
titles("Figure 10. Box Plot: Median and Interquartile Range of Vital Signs by Treatment Arm",
bold = TRUE, font_size = 12, align = "left") |>
page_by(VSTESTCD, label = "Lab Test: ", format = lbfmt, blank_row = "none") |>
footnotes(
"Source: example6.rtf. {version$version.string}",
"Note: Boxes span the interquartile range (25th to 75th percentile); horizontal line = median;",
"whiskers = 1.5×IQR; individual outliers are those beyond this range.",
font_size = 9, italics = TRUE, blank_row = "none"
)
put("Create report output path")
pth <- file.path(tempdir(), "example6.rtf")
put("Create report")
rpt <- create_report(pth, font = "Arial", font_size = 10, output_type = "RTF") |>
page_header("Sponsor: Company", right = "Study: ABC", blank_row = "below") |>
add_content(plt) |>
page_footer("Date Produced: {fapply(Sys.Date(), 'date7')}", right = "Page [pg] of [tpg]")
put("Write report to file system")
write_report(rpt)
put("Close log")
log_close()
# View report
# file.show(pth)
# View log
# file.show(lgpth)Log
Here is the log for the above program:
=========================================================================
Log Path: C:/Users/dbosa/AppData/Local/Temp/Rtmpq0yZA5/log/example6.log
Program Path: C:/Studies/Testing1/P6b.R
Working Directory: C:/Studies/Testing1
User Name: dbosa
R Version: 4.4.3 (2025-02-28 ucrt)
Machine: SOCRATES x86-64
Operating System: Windows 10 x64 build 26100
Base Packages: stats graphics grDevices utils datasets methods base
Other Packages: tidylog_1.1.0 ggplot2_3.5.1 procs_1.0.7 reporter_1.4.5
libr_1.3.9 logr_1.3.9 fmtr_1.7.0 common_1.1.4 sassy_1.2.9
Log Start Time: 2025-12-12 14:27:32.328372
=========================================================================
=========================================================================
Prepare Data
=========================================================================
Load data files
# library 'dat': 2 items
- attributes: csv not loaded
- path: C:/Users/dbosa/AppData/Local/R/win-library/4.4/sassy/extdata
- items:
Name Extension Rows Cols Size LastModified
1 DM csv 87 24 45.8 Kb 2025-12-12 08:37:33
2 VS csv 3358 17 467.7 Kb 2025-12-12 08:37:33
Prepare factor levels
SCREENING
DAY 1
WEEK 2
WEEK 4
WEEK 6
WEEK 8
WEEK 12
WEEK 16
ARM A
ARM B
SYSBP
DIABP
PULSE
RESP
Prepare data for analysis
datastep: columns decreased from 24 to 6
# A tibble: 1,231 × 6
STUDYID USUBJID ARM VISIT VSTESTCD VSORRES
<chr> <chr> <fct> <fct> <fct> <dbl>
1 ABC ABC-01-050 ARM B SCREENING DIABP 80
2 ABC ABC-01-050 ARM B DAY 1 DIABP 78
3 ABC ABC-01-050 ARM B WEEK 2 DIABP 64
4 ABC ABC-01-050 ARM B WEEK 4 DIABP 86
5 ABC ABC-01-050 ARM B WEEK 6 DIABP 70
6 ABC ABC-01-050 ARM B WEEK 8 DIABP 80
7 ABC ABC-01-050 ARM B WEEK 12 DIABP 64
8 ABC ABC-01-050 ARM B WEEK 16 DIABP 82
9 ABC ABC-01-050 ARM B SCREENING PULSE 76
10 ABC ABC-01-050 ARM B DAY 1 PULSE 68
# ℹ 1,221 more rows
# ℹ Use `print(n = ...)` to see more rows
=========================================================================
Create Plot
=========================================================================
Assign Colors
Define Boxplot
Create format for lab codes
# A user-defined format: 4 conditions
Name Type Expression Label Order
1 obj U x == "SYSBP" Systolic Blood Pressure (mmHg) NA
2 obj U x == "DIABP" Diastolic Blood Pressure (mmHg) NA
3 obj U x == "PULSE" Pulse (bpm) NA
4 obj U x == "RESP" Respirations (bpm) NA
=========================================================================
Report
=========================================================================
Create plot object definition
Create report output path
Create report
Write report to file system
# A report specification: 4 pages
- file_path: 'C:\Users\dbosa\AppData\Local\Temp\Rtmpq0yZA5/example6.rtf'
- output_type: RTF
- units: inches
- orientation: landscape
- margins: top 0.5 bottom 0.5 left 1 right 1
- line size/count: 9/42
- page_header: left=Sponsor: Company right=Study: ABC
- page_footer: left=Date Produced: 12DEC25 center= right=Page [pg] of [tpg]
- content:
# A plot specification:
- data: 1231 rows, 6 cols
- layers: 1
- height: 4
- width: 7
- page by: VSTESTCD
- title 1: 'Figure 10. Box Plot: Median and Interquartile Range of Vital Signs by Treatment Arm'
- footnote 1: 'Source: example6.rtf. R version 4.4.3 (2025-02-28 ucrt)'
- footnote 2: 'Note: Boxes span the interquartile range (25th to 75th percentile); horizontal line = median;'
- footnote 3: 'whiskers = 1.5×IQR; individual outliers are those beyond this range.'
Close log
=========================================================================
Log End Time: 2025-12-12 14:27:40.667776
Log Elapsed Time: 0 00:00:08
=========================================================================