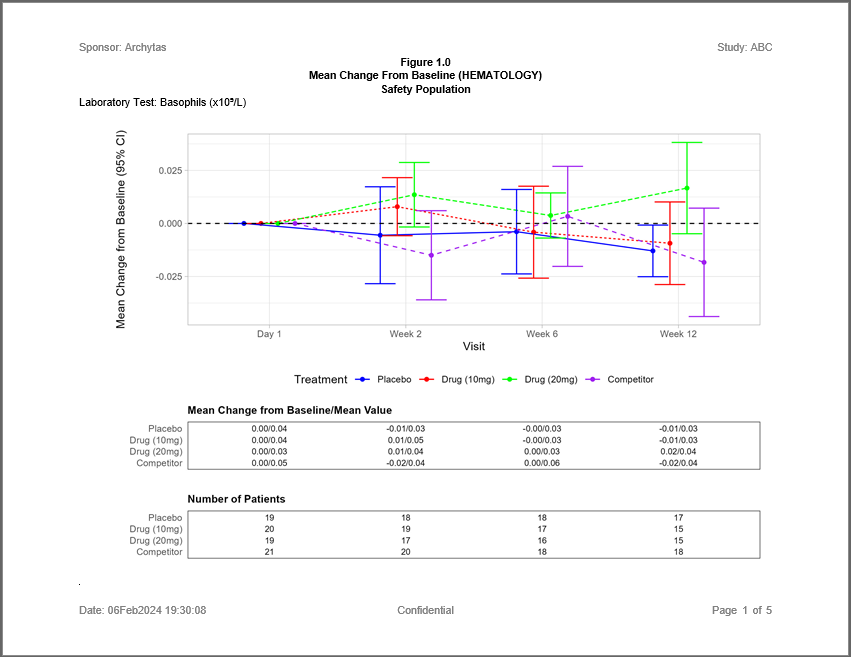
The following example shows a Mean Change from Baseline chart for Hematology Lab Values with supporting tables for the mean value and number of patients. The figure is accomplished using the ggplot and patchwork packages, and data preparation that features the procs package.
Program
Note the following about this example:
- The procs package calculates mean values and confidence intervals for each treatment group and visit.
- The ggplot2 package can create small tables in addition to charts.
- The patchwork
package can combine the ggplot chart and table into a single
image.
- The patchwork images are dynamically added to a report object from the reporter package. Titles are customized for each page.
library(sassy)
library(ggplot2)
library(patchwork)
options("logr.notes" = FALSE,
"logr.autolog" = TRUE,
"procs.print" = FALSE)
# Get temp location for log and report output
tmp <- tempdir()
lf <- log_open(file.path(tmp, "example13.log"))
# Get data ----------------------------------------------------------------
sep("Get data")
# Get sample data path
pth <- system.file("extdata", package = "sassy")
put("Open data library")
libname(sdtm, pth, "csv")
# Create Formats ----------------------------------------------------------
sep("Create Formats")
put("Format for visits")
vfmt <- value(condition(x == "DAY 1", "Day 1"),
condition(x == "WEEK 2", "Week 2"),
condition(x == "WEEK 4", "Week 4"),
condition(x == "WEEK 6", "Week 6"),
condition(x == "WEEK 12", "Week 12"),
as.factor = TRUE)
put("Format for ARMs")
afmt <- value(condition(x == "ARM A", "Placebo"),
condition(x == "ARM B", "Drug (10mg)"),
condition(x == "ARM C", "Drug (20mg)"),
condition(x == "ARM D", "Competitor"),
as.factor = TRUE)
# Prepare data ------------------------------------------------------------
sep("Prepare data")
put("Pull out needed visits and columns")
lbsub1 <- subset(sdtm$LB, VISIT %in% toupper(levels(vfmt)),
v(USUBJID, VISIT, VISITNUM, LBORRES, LBCAT, LBORRESU, LBTEST,
LBTESTCD, LBBLFL)) |> put()
put("Pull out baseline subset")
lbsub2 <- subset(lbsub1, LBBLFL == 'Y',
v(USUBJID, VISIT, LBORRES, LBCAT, LBTESTCD)) |> put()
put("Merge and calculate change from baseline")
datastep(lbsub1, merge = lbsub2, merge_by = v(USUBJID, LBCAT, LBTESTCD),
rename = v(LBORRES.1 = LBORRES, LBORRES.2 = BLBORES, VISIT.1 = VISIT),
drop = VISIT.2, {
# Convert to double
LBORRES.1 <- suppressWarnings(as.double(LBORRES.1))
# Convert to double
LBORRES.2 <- suppressWarnings(as.double(LBORRES.2))
# Calculate Change from baseline
if (!(is.na(LBORRES.1) | is.na(LBORRES.2))) {
LBCHG <- LBORRES.1 - LBORRES.2
} else {
LBCHG <- NA
}
}) -> lbsub
put("Pull needed ARMs and columns for DM")
dmsub <- subset(sdtm$DM, ARM != "SCREEN FAILURE", v(USUBJID, ARMCD, ARM)) |> put()
put("Merge DM with LB to get subject treatments")
datastep(lbsub, merge = dmsub, merge_by = USUBJID,
where = expression(toupper(VISIT) != 'SCREENING'),
{
VISIT <- fapply(VISIT, vfmt)
}) -> lbdat
# Prepare lab test labels -------------------------------------------------
sep("Lab test labels")
put("Get lookup data for lab tests")
proc_sort(lbdat, by = v(LBTESTCD, LBTEST, LBORRESU),
keep = v(LBTESTCD, LBTEST, LBORRESU),
options = nodupkey) -> tcodes
put("Create test label with units")
datastep(tcodes, where = expression(is.na(LBORRESU) == FALSE),
keep = v(LBTESTCD, LABEL),
{
LABEL <- paste0(LBTEST, " (", LBORRESU, ")")
}) -> tfmtdat
put("Create lab value lookup")
tfmt <- tfmtdat$LABEL
names(tfmt) <- tfmtdat$LBTESTCD
put("Apply superscripts as needed")
tfmt <- sub("(9)", supsc('9'), tfmt, fixed = TRUE)
tfmt <- sub("(12)", supsc('12'), tfmt, fixed = TRUE)
# Calculate statistics ----------------------------------------------------
sep("Calculate statistics")
put("Get statistics for change from baseline")
proc_means(lbdat, by = LBTESTCD,
class = v(ARM, VISIT),
var = LBCHG,
stats = v(n, mean, std, clm),
options = nway) -> datcl
put("Get statistics for mean lab value")
proc_means(lbdat, by = LBTESTCD,
class = v(ARM, VISIT),
var = LBORRES,
stats = v(n, mean),
options = nway) -> datmn
put("Add mean lab values to change from baseline")
datcl$MEANV <- datmn$MEAN
put("Apply formats")
datcl$BY <- fapply(datcl$BY, tfmt)
datcl$CLASS1 <- fapply(datcl$CLASS1, afmt)
# Create report -----------------------------------------------------------
sep("Create report")
put("Create output path")
pth <- file.path(tmp, "output/example13.rtf") |> put()
put("Create report first so content can be added dynamically")
rpt <- create_report(pth, output_type = "RTF",
font = "Arial", font_size = 10) |>
page_header("Sponsor: Archytas", right = "Study: ABC") |>
titles("Figure 1.0", "Mean Change From Baseline (HEMATOLOGY)", "Safety Population",
bold = TRUE, blank_row = "none") |>
page_footer("Date: " %p% fapply(Sys.time(), "%d%b%Y %H:%M:%S"),
"Confidential", "Page [pg] of [tpg]")
put("Loop through lab tests")
for (tst in unique(datcl$BY)) {
put("**** Lab Test: " %p% tst %p% " ****")
put("Apply subset")
dat <- subset(datcl, BY == tst)
put("Create plot")
p <- ggplot(data = dat, aes(x = CLASS2, y = MEAN, group = CLASS1)) +
geom_point(aes(color = CLASS1), size = 1.5, position = position_dodge(width = 0.5)) +
geom_line(aes(color = CLASS1, linetype = CLASS1), position = position_dodge(width = 0.5)) +
geom_errorbar(aes(x = CLASS2, ymin = LCLM, ymax = UCLM, color = CLASS1),
position = position_dodge(width = 0.5)) +
theme_light() +
scale_linetype(guide = "none") +
scale_color_manual(values = c("blue", "red", "green", "purple")) +
theme(legend.position = "bottom", plot.title = element_text(size = 11, hjust = 0)) +
guides(color = guide_legend(title = "Treatment")) +
labs(x = 'Visit', y = '\n\nMean Change from Baseline (95% CI)') +
geom_hline(yintercept = 0, linetype = 'dashed')
put("Create table for mean change/mean value")
t1 <- ggplot(data = dat) +
geom_text(aes(CLASS1, x = CLASS2, label = fapply2(MEAN, MEANV, "%4.2f", "%.2f", sep = "/"),
hjust = 0.5, vjust = 0.5), size = 8 / .pt) +
ggtitle("Mean Change from Baseline/Mean Value") +
scale_y_discrete(limits = rev) +
theme_bw() +
theme(
axis.line = element_blank(),
panel.grid = element_blank(),
axis.ticks = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank(),
axis.text.x = element_text(color = "white"),
plot.title = element_text(size =10, hjust = 0, face = "bold")
)
put("Create table for patient counts")
t2 <- ggplot(data = dat) +
geom_text(aes(CLASS1, x = CLASS2, label = as.character(N),
hjust = 0.5, vjust = 0.5), size = 8 / .pt) +
ggtitle("Number of Patients") +
scale_y_discrete(limits = rev) +
theme_bw() +
theme(
axis.line = element_blank(),
panel.grid = element_blank(),
axis.ticks = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank(),
axis.text.x = element_text(color = "white"),
plot.title = element_text(size = 10, hjust = 0, face = "bold")
)
put("Patch together plot and tables")
plts <- p + t1 + t2 + plot_layout(ncol = 1, nrow = 3,
widths = c(1, 2, 2), heights = c(8, 2, 2))
put("Create plot content")
plt1 <- create_plot(plts, height = 6, width = 9) |>
titles("Laboratory Test: " %p% tst, align = "left", blank_row = "below")
put("Add content to report")
rpt <- rpt |>
add_content(plt1, page_break = TRUE, blank_row = "none")
}
put("Write out report to file system")
res <- write_report(rpt)
# Clean Up ----------------------------------------------------------------
sep("Clean Up")
log_close()
# View report
# file.show(res$modified_path)
# View log
# file.show(lf)Log
Here is the log produced by the above mean change from baseline example:
=========================================================================
Log Path: C:/Users/dbosa/AppData/Local/Temp/RtmpuCtffJ/log/example13.log
Program Path: C:/packages/Testing/sassytests/ControlChart.R
Working Directory: C:/packages/Testing
User Name: dbosa
R Version: 4.3.2 (2023-10-31 ucrt)
Machine: SOCRATES x86-64
Operating System: Windows 10 x64 build 22621
Base Packages: stats graphics grDevices utils datasets methods base Other
Packages: tidylog_1.0.2 patchwork_1.1.3 ggplot2_3.4.4 procs_1.0.5
reporter_1.4.4 libr_1.2.9 logr_1.3.5 fmtr_1.6.2 common_1.1.1 sassy_1.2.1
Log Start Time: 2024-02-02 22:44:16.986801
=========================================================================
=========================================================================
Get data
=========================================================================
Open data library
# library 'sdtm': 15 items
- attributes: csv not loaded
- path: ./data/abc/SDTM
- items:
Name Extension Rows Cols Size LastModified
1 AE csv 150 27 88.3 Kb 2020-09-18 14:30:23
2 DA csv 3587 18 528 Kb 2020-09-18 14:30:23
3 DM csv 87 24 45.3 Kb 2020-09-18 14:30:23
4 DS csv 174 9 33.9 Kb 2020-09-18 14:30:23
5 DS_IHOR csv 174 9 33.9 Kb 2020-09-18 14:30:23
6 EX csv 84 11 26.2 Kb 2020-09-18 14:30:23
7 IE csv 2 14 13.2 Kb 2020-09-18 14:30:23
8 LB csv 2069 12 211.2 Kb 2024-02-02 22:42:43
9 LB4 csv 23607 27 5.4 Mb 2020-09-18 14:30:23
10 LB6 csv 7810 12 879 Kb 2024-02-01 00:20:30
11 PE csv 1854 17 275.7 Kb 2020-09-18 14:30:24
12 QS csv 13316 17 1.7 Mb 2020-09-18 14:30:24
13 SUPPEX csv 639 10 63.8 Kb 2020-09-18 14:30:24
14 SV csv 685 10 70.1 Kb 2020-09-18 14:30:24
15 VS csv 3358 17 467.2 Kb 2020-09-18 14:30:24
=========================================================================
Create Formats
=========================================================================
Format for visits
# A user-defined format: 5 conditions
- as.factor: TRUE
Name Type Expression Label Order
1 obj U x == "DAY 1" Day 1 NA
2 obj U x == "WEEK 2" Week 2 NA
3 obj U x == "WEEK 4" Week 4 NA
4 obj U x == "WEEK 6" Week 6 NA
5 obj U x == "WEEK 12" Week 12 NA
Format for ARMs
# A user-defined format: 4 conditions
- as.factor: TRUE
Name Type Expression Label Order
1 obj U x == "ARM A" Placebo NA
2 obj U x == "ARM B" Drug (10mg) NA
3 obj U x == "ARM C" Drug (20mg) NA
4 obj U x == "ARM D" Competitor NA
=========================================================================
Prepare data
=========================================================================
Pull out needed visits and columns
# A tibble: 1,578 × 9
USUBJID VISIT VISITNUM LBORRES LBCAT LBORRESU LBTEST LBTESTCD LBBLFL
<chr> <chr> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr>
1 ABC-01-049 DAY 1 1 0.04 HEMATOLOGY x10(9)/L Basophils BASO Y
2 ABC-01-049 WEEK 2 2 0.01 HEMATOLOGY x10(9)/L Basophils BASO <NA>
3 ABC-01-049 WEEK 6 6 0.06 HEMATOLOGY x10(9)/L Basophils BASO <NA>
4 ABC-01-049 WEEK 12 12 0.03 HEMATOLOGY x10(9)/L Basophils BASO <NA>
5 ABC-01-049 DAY 1 1 0.19 HEMATOLOGY x10(9)/L Eosinophils EOS Y
6 ABC-01-049 WEEK 2 2 0.19 HEMATOLOGY x10(9)/L Eosinophils EOS <NA>
7 ABC-01-049 WEEK 6 6 0.21 HEMATOLOGY x10(9)/L Eosinophils EOS <NA>
8 ABC-01-049 WEEK 12 12 0.18 HEMATOLOGY x10(9)/L Eosinophils EOS <NA>
9 ABC-01-049 DAY 1 1 42.6 HEMATOLOGY % Hematocrit HCT Y
10 ABC-01-049 WEEK 2 2 44.3 HEMATOLOGY % Hematocrit HCT <NA>
# ℹ 1,568 more rows
# ℹ Use `print(n = ...)` to see more rows
Pull out baseline subset
# A tibble: 409 × 5
USUBJID VISIT LBORRES LBCAT LBTESTCD
<chr> <chr> <dbl> <chr> <chr>
1 ABC-01-049 DAY 1 0.04 HEMATOLOGY BASO
2 ABC-01-049 DAY 1 0.19 HEMATOLOGY EOS
3 ABC-01-049 DAY 1 42.6 HEMATOLOGY HCT
4 ABC-01-049 DAY 1 15 HEMATOLOGY HGB
5 ABC-01-049 DAY 1 1.91 HEMATOLOGY LYM
6 ABC-01-050 DAY 1 0.04 HEMATOLOGY BASO
7 ABC-01-050 DAY 1 0.33 HEMATOLOGY EOS
8 ABC-01-050 DAY 1 44.6 HEMATOLOGY HCT
9 ABC-01-050 DAY 1 15.7 HEMATOLOGY HGB
10 ABC-01-050 DAY 1 1.23 HEMATOLOGY LYM
# ℹ 399 more rows
# ℹ Use `print(n = ...)` to see more rows
Merge and calculate change from baseline
datastep: columns increased from 9 to 11
# A tibble: 1,578 × 11
USUBJID VISIT VISITNUM LBORRES LBCAT LBORRESU LBTEST LBTESTCD LBBLFL BLBORES
<chr> <chr> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <dbl>
1 ABC-01-049 WEEK 12 12 0.03 HEMATO… x10(9)/L Basop… BASO <NA> 0.04
2 ABC-01-049 WEEK 2 2 0.01 HEMATO… x10(9)/L Basop… BASO <NA> 0.04
3 ABC-01-049 DAY 1 1 0.04 HEMATO… x10(9)/L Basop… BASO Y 0.04
4 ABC-01-049 WEEK 6 6 0.06 HEMATO… x10(9)/L Basop… BASO <NA> 0.04
5 ABC-01-049 WEEK 12 12 0.18 HEMATO… x10(9)/L Eosin… EOS <NA> 0.19
6 ABC-01-049 DAY 1 1 0.19 HEMATO… x10(9)/L Eosin… EOS Y 0.19
7 ABC-01-049 WEEK 2 2 0.19 HEMATO… x10(9)/L Eosin… EOS <NA> 0.19
8 ABC-01-049 WEEK 6 6 0.21 HEMATO… x10(9)/L Eosin… EOS <NA> 0.19
9 ABC-01-049 WEEK 12 12 45.3 HEMATO… % Hemat… HCT <NA> 42.6
10 ABC-01-049 WEEK 2 2 44.3 HEMATO… % Hemat… HCT <NA> 42.6
# ℹ 1,568 more rows
# ℹ 1 more variable: LBCHG <dbl>
# ℹ Use `print(n = ...)` to see more rows
Pull needed ARMs and columns for DM
# A tibble: 85 × 3
USUBJID ARMCD ARM
<chr> <chr> <chr>
1 ABC-01-049 4 ARM D
2 ABC-01-050 2 ARM B
3 ABC-01-051 1 ARM A
4 ABC-01-052 3 ARM C
5 ABC-01-053 2 ARM B
6 ABC-01-054 4 ARM D
7 ABC-01-055 3 ARM C
8 ABC-01-056 1 ARM A
9 ABC-01-113 4 ARM D
10 ABC-01-114 2 ARM B
# ℹ 75 more rows
# ℹ Use `print(n = ...)` to see more rows
Merge DM with LB to get subject treatments
datastep: columns increased from 11 to 13
# A tibble: 1,578 × 13
USUBJID VISIT VISITNUM LBORRES LBCAT LBORRESU LBTEST LBTESTCD LBBLFL BLBORES
<chr> <ord> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <dbl>
1 ABC-01-049 Week 12 12 2.59 HEMATO… x10(9)/L Lymph… LYM <NA> 1.91
2 ABC-01-049 Week 6 6 2.35 HEMATO… x10(9)/L Lymph… LYM <NA> 1.91
3 ABC-01-049 Week 2 2 2.59 HEMATO… x10(9)/L Lymph… LYM <NA> 1.91
4 ABC-01-049 Week 6 6 0.06 HEMATO… x10(9)/L Basop… BASO <NA> 0.04
5 ABC-01-049 Week 12 12 0.18 HEMATO… x10(9)/L Eosin… EOS <NA> 0.19
6 ABC-01-049 Week 12 12 15.9 HEMATO… g/dL Hemog… HGB <NA> 15
7 ABC-01-049 Week 2 2 0.19 HEMATO… x10(9)/L Eosin… EOS <NA> 0.19
8 ABC-01-049 Week 6 6 0.21 HEMATO… x10(9)/L Eosin… EOS <NA> 0.19
9 ABC-01-049 Week 12 12 45.3 HEMATO… % Hemat… HCT <NA> 42.6
10 ABC-01-049 Day 1 1 1.91 HEMATO… x10(9)/L Lymph… LYM Y 1.91
# ℹ 1,568 more rows
# ℹ 3 more variables: LBCHG <dbl>, ARMCD <chr>, ARM <chr>
# ℹ Use `print(n = ...)` to see more rows
=========================================================================
Lab test labels
=========================================================================
Get lookup data for lab tests
proc_sort: input data set 10 rows and 13 columns
by: LBTESTCD LBTEST LBORRESU
keep: LBTESTCD LBTEST LBORRESU
order: a a a
options: nodupkey
output data set 10 rows and 3 columns
# A tibble: 10 × 3
LBTESTCD LBTEST LBORRESU
<chr> <chr> <chr>
1 BASO Basophils x10(9)/L
2 BASO Basophils <NA>
3 EOS Eosinophils x10(9)/L
4 EOS Eosinophils <NA>
5 HCT Hematocrit %
6 HCT Hematocrit <NA>
7 HGB Hemoglobin g/dL
8 HGB Hemoglobin <NA>
9 LYM Lymphocytes x10(9)/L
10 LYM Lymphocytes <NA>
Create test label with units
datastep: columns decreased from 3 to 2
# A tibble: 5 × 2
LBTESTCD LABEL
<chr> <chr>
1 BASO Basophils (x10(9)/L)
2 EOS Eosinophils (x10(9)/L)
3 HCT Hematocrit (%)
4 HGB Hemoglobin (g/dL)
5 LYM Lymphocytes (x10(9)/L)
Create lab value lookup
Apply superscripts as needed
=========================================================================
Calculate statistics
=========================================================================
Get statistics for change from baseline
proc_means: input data set 1578 rows and 13 columns
by: LBTESTCD
class: ARM VISIT
var: LBCHG
stats: n mean std clm
view: TRUE
output: 1 datasets
CLASS1 CLASS2 BY TYPE FREQ VAR N MEAN STD LCLM
1 ARM A Day 1 BASO 3 19 LBCHG 19 0.000000e+00 0.00000000 0.000000000
2 ARM A Week 2 BASO 3 19 LBCHG 18 -5.555556e-03 0.04591837 -0.028390224
3 ARM A Week 6 BASO 3 19 LBCHG 18 -3.888889e-03 0.04002042 -0.023790575
4 ARM A Week 12 BASO 3 18 LBCHG 17 -1.294118e-02 0.02365500 -0.025103453
5 ARM B Day 1 BASO 3 20 LBCHG 20 0.000000e+00 0.00000000 0.000000000
6 ARM B Week 2 BASO 3 20 LBCHG 19 7.894737e-03 0.02839776 -0.005792545
7 ARM B Week 6 BASO 3 18 LBCHG 17 -4.117647e-03 0.04213947 -0.025783766
8 ARM B Week 12 BASO 3 18 LBCHG 15 -9.333333e-03 0.03514595 -0.028796514
9 ARM C Day 1 BASO 3 19 LBCHG 19 0.000000e+00 0.00000000 0.000000000
10 ARM C Week 2 BASO 3 18 LBCHG 17 1.352941e-02 0.02956797 -0.001673034
11 ARM C Week 6 BASO 3 17 LBCHG 16 3.750000e-03 0.01995829 -0.006885022
12 ARM C Week 12 BASO 3 15 LBCHG 15 1.666667e-02 0.03885259 -0.004849181
13 ARM D Day 1 BASO 3 25 LBCHG 21 0.000000e+00 0.00000000 0.000000000
14 ARM D Week 2 BASO 3 22 LBCHG 20 -1.500000e-02 0.04489754 -0.036012697
15 ARM D Week 6 BASO 3 19 LBCHG 18 3.333333e-03 0.04740315 -0.020239700
16 ARM D Week 12 BASO 3 20 LBCHG 18 -1.833333e-02 0.05136375 -0.043875928
17 ARM A Day 1 EOS 3 19 LBCHG 19 0.000000e+00 0.00000000 0.000000000
18 ARM A Week 2 EOS 3 19 LBCHG 18 8.888889e-03 0.09177267 -0.036748584
19 ARM A Week 6 EOS 3 19 LBCHG 18 -5.555556e-04 0.11132740 -0.055917371
20 ARM A Week 12 EOS 3 18 LBCHG 17 -2.941176e-03 0.10263800 -0.055712766
21 ARM B Day 1 EOS 3 20 LBCHG 20 0.000000e+00 0.00000000 0.000000000
22 ARM B Week 2 EOS 3 20 LBCHG 19 -2.052632e-02 0.06875671 -0.053665990
23 ARM B Week 6 EOS 3 18 LBCHG 17 -7.058824e-03 0.06687345 -0.041441981
24 ARM B Week 12 EOS 3 18 LBCHG 15 -2.333333e-02 0.05924123 -0.056140035
25 ARM C Day 1 EOS 3 19 LBCHG 19 0.000000e+00 0.00000000 0.000000000
26 ARM C Week 2 EOS 3 18 LBCHG 17 -3.470588e-02 0.06549023 -0.068377853
27 ARM C Week 6 EOS 3 17 LBCHG 16 -2.875000e-02 0.07237633 -0.067316625
28 ARM C Week 12 EOS 3 15 LBCHG 15 -2.533333e-02 0.10020455 -0.080824765
29 ARM D Day 1 EOS 3 25 LBCHG 21 0.000000e+00 0.00000000 0.000000000
30 ARM D Week 2 EOS 3 22 LBCHG 20 4.000000e-03 0.09150209 -0.038824294
31 ARM D Week 6 EOS 3 19 LBCHG 18 -1.388889e-02 0.09437964 -0.060822779
32 ARM D Week 12 EOS 3 20 LBCHG 18 -3.111111e-02 0.13437826 -0.097935847
33 ARM A Day 1 HCT 3 20 LBCHG 20 0.000000e+00 0.00000000 0.000000000
34 ARM A Week 2 HCT 3 20 LBCHG 20 3.400000e-01 1.67438913 -0.443638233
35 ARM A Week 6 HCT 3 19 LBCHG 19 4.736842e-02 2.10353365 -0.966502342
36 ARM A Week 12 HCT 3 19 LBCHG 19 -1.473684e-01 2.20185993 -1.208630934
37 ARM B Day 1 HCT 3 21 LBCHG 21 0.000000e+00 0.00000000 0.000000000
38 ARM B Week 2 HCT 3 20 LBCHG 20 5.150000e-01 1.67905832 -0.270823483
39 ARM B Week 6 HCT 3 18 LBCHG 18 -4.000000e-01 2.17147280 -1.479848018
40 ARM B Week 12 HCT 3 18 LBCHG 17 -1.941176e-01 1.86395768 -1.152476254
41 ARM C Day 1 HCT 3 19 LBCHG 19 0.000000e+00 0.00000000 0.000000000
42 ARM C Week 2 HCT 3 18 LBCHG 17 -8.823529e-02 1.87078939 -1.050106442
43 ARM C Week 6 HCT 3 17 LBCHG 16 -3.000000e-01 1.78848167 -1.253014608
44 ARM C Week 12 HCT 3 16 LBCHG 15 -6.066667e-01 2.42147141 -1.947632839
45 ARM D Day 1 HCT 3 23 LBCHG 21 0.000000e+00 0.00000000 0.000000000
46 ARM D Week 2 HCT 3 22 LBCHG 20 1.900000e-01 1.74654546 -0.627408438
47 ARM D Week 6 HCT 3 20 LBCHG 18 7.894799e-16 1.35559841 -0.674123232
48 ARM D Week 12 HCT 3 20 LBCHG 18 -5.833333e-01 1.52286495 -1.340636255
49 ARM A Day 1 HGB 3 20 LBCHG 20 0.000000e+00 0.00000000 0.000000000
50 ARM A Week 2 HGB 3 20 LBCHG 20 7.000000e-02 0.49957877 -0.163810061
51 ARM A Week 6 HGB 3 19 LBCHG 19 2.105263e-02 0.63646502 -0.285713688
52 ARM A Week 12 HGB 3 19 LBCHG 19 -1.210526e-01 0.65453881 -0.436530239
53 ARM B Day 1 HGB 3 21 LBCHG 21 0.000000e+00 0.00000000 0.000000000
54 ARM B Week 2 HGB 3 20 LBCHG 20 1.300000e-01 0.56110793 -0.132606593
55 ARM B Week 6 HGB 3 18 LBCHG 18 -8.333333e-02 0.74379789 -0.453215320
56 ARM B Week 12 HGB 3 18 LBCHG 17 -3.529412e-02 0.54651301 -0.316285179
57 ARM C Day 1 HGB 3 19 LBCHG 19 0.000000e+00 0.00000000 0.000000000
58 ARM C Week 2 HGB 3 18 LBCHG 17 0.000000e+00 0.65383484 -0.336170855
59 ARM C Week 6 HGB 3 17 LBCHG 16 -8.750000e-02 0.61196405 -0.413592625
60 ARM C Week 12 HGB 3 16 LBCHG 15 -1.666667e-01 0.64438971 -0.523517792
61 ARM D Day 1 HGB 3 23 LBCHG 21 0.000000e+00 0.00000000 0.000000000
62 ARM D Week 2 HGB 3 22 LBCHG 20 -2.500000e-02 0.55995771 -0.287068273
63 ARM D Week 6 HGB 3 20 LBCHG 18 -1.111111e-02 0.42962212 -0.224757164
64 ARM D Week 12 HGB 3 20 LBCHG 18 -2.000000e-01 0.51449576 -0.455852720
65 ARM A Day 1 LYM 3 19 LBCHG 19 0.000000e+00 0.00000000 0.000000000
66 ARM A Week 2 LYM 3 19 LBCHG 18 6.166667e-02 0.44072467 -0.157500556
67 ARM A Week 6 LYM 3 19 LBCHG 18 -5.555556e-03 0.47330157 -0.240922891
68 ARM A Week 12 LYM 3 18 LBCHG 17 -3.529412e-03 0.42161803 -0.220305401
69 ARM B Day 1 LYM 3 20 LBCHG 20 0.000000e+00 0.00000000 0.000000000
70 ARM B Week 2 LYM 3 20 LBCHG 19 2.278947e-01 0.44226160 0.014731470
71 ARM B Week 6 LYM 3 18 LBCHG 17 5.588235e-02 0.19624789 -0.045019003
72 ARM B Week 12 LYM 3 18 LBCHG 15 1.166667e-01 0.30513853 -0.052313420
73 ARM C Day 1 LYM 3 19 LBCHG 19 0.000000e+00 0.00000000 0.000000000
74 ARM C Week 2 LYM 3 18 LBCHG 17 6.117647e-02 0.25458010 -0.069716541
75 ARM C Week 6 LYM 3 17 LBCHG 16 -3.062500e-02 0.29862951 -0.189753433
76 ARM C Week 12 LYM 3 15 LBCHG 15 -2.133333e-02 0.25939123 -0.164979408
77 ARM D Day 1 LYM 3 25 LBCHG 21 0.000000e+00 0.00000000 0.000000000
78 ARM D Week 2 LYM 3 22 LBCHG 20 -6.450000e-02 0.53546315 -0.315104470
79 ARM D Week 6 LYM 3 19 LBCHG 18 -4.333333e-02 0.43414554 -0.259228833
80 ARM D Week 12 LYM 3 20 LBCHG 18 -1.461111e-01 0.41560925 -0.352788727
UCLM
1 0.0000000000
2 0.0172791128
3 0.0160127976
4 -0.0007788999
5 0.0000000000
6 0.0215820188
7 0.0175484714
8 0.0101298474
9 0.0000000000
10 0.0287318579
11 0.0143850220
12 0.0381825143
13 0.0000000000
14 0.0060126975
15 0.0269063663
16 0.0072092609
17 0.0000000000
18 0.0545263620
19 0.0548062599
20 0.0498304131
21 0.0000000000
22 0.0126133584
23 0.0273243337
24 0.0094733686
25 0.0000000000
26 -0.0010339117
27 0.0098166246
28 0.0301580983
29 0.0000000000
30 0.0468242940
31 0.0330450016
32 0.0357136247
33 0.0000000000
34 1.1236382332
35 1.0612391846
36 0.9138940923
37 0.0000000000
38 1.3008234830
39 0.6798480184
40 0.7642409602
41 0.0000000000
42 0.8736358538
43 0.6530146081
44 0.7342995059
45 0.0000000000
46 1.0074084379
47 0.6741232315
48 0.1739695884
49 0.0000000000
50 0.3038100615
51 0.3278189507
52 0.1944249760
53 0.0000000000
54 0.3926065932
55 0.2865486531
56 0.2456969440
57 0.0000000000
58 0.3361708555
59 0.2385926247
60 0.1901844590
61 0.0000000000
62 0.2370682730
63 0.2025349414
64 0.0558527199
65 0.0000000000
66 0.2808338892
67 0.2298117794
68 0.2132465772
69 0.0000000000
70 0.4410580040
71 0.1567837086
72 0.2856467534
73 0.0000000000
74 0.1920694820
75 0.1285034325
76 0.1223127412
77 0.0000000000
78 0.1861044703
79 0.1725621663
80 0.0605665048
Get statistics for mean lab value
proc_means: input data set 1578 rows and 13 columns
by: LBTESTCD
class: ARM VISIT
var: LBORRES
stats: n mean
view: TRUE
output: 1 datasets
CLASS1 CLASS2 BY TYPE FREQ VAR N MEAN
1 ARM A Day 1 BASO 3 19 LBORRES 19 0.03789474
2 ARM A Week 2 BASO 3 19 LBORRES 19 0.03105263
3 ARM A Week 6 BASO 3 19 LBORRES 19 0.03473684
4 ARM A Week 12 BASO 3 18 LBORRES 18 0.02666667
5 ARM B Day 1 BASO 3 20 LBORRES 20 0.04100000
6 ARM B Week 2 BASO 3 20 LBORRES 20 0.04800000
7 ARM B Week 6 BASO 3 18 LBORRES 18 0.03333333
8 ARM B Week 12 BASO 3 18 LBORRES 16 0.03250000
9 ARM C Day 1 BASO 3 19 LBORRES 19 0.02684211
10 ARM C Week 2 BASO 3 18 LBORRES 18 0.03944444
11 ARM C Week 6 BASO 3 17 LBORRES 17 0.03117647
12 ARM C Week 12 BASO 3 15 LBORRES 15 0.04466667
13 ARM D Day 1 BASO 3 25 LBORRES 21 0.05333333
14 ARM D Week 2 BASO 3 22 LBORRES 22 0.03863636
15 ARM D Week 6 BASO 3 19 LBORRES 19 0.06210526
16 ARM D Week 12 BASO 3 20 LBORRES 20 0.03950000
17 ARM A Day 1 EOS 3 19 LBORRES 19 0.15947368
18 ARM A Week 2 EOS 3 19 LBORRES 19 0.16631579
19 ARM A Week 6 EOS 3 19 LBORRES 19 0.16578947
20 ARM A Week 12 EOS 3 18 LBORRES 18 0.17722222
21 ARM B Day 1 EOS 3 20 LBORRES 20 0.22000000
22 ARM B Week 2 EOS 3 20 LBORRES 20 0.19600000
23 ARM B Week 6 EOS 3 18 LBORRES 18 0.21722222
24 ARM B Week 12 EOS 3 18 LBORRES 16 0.20250000
25 ARM C Day 1 EOS 3 19 LBORRES 19 0.20315789
26 ARM C Week 2 EOS 3 18 LBORRES 18 0.17944444
27 ARM C Week 6 EOS 3 17 LBORRES 17 0.18352941
28 ARM C Week 12 EOS 3 15 LBORRES 15 0.18800000
29 ARM D Day 1 EOS 3 25 LBORRES 21 0.29095238
30 ARM D Week 2 EOS 3 22 LBORRES 22 0.25681818
31 ARM D Week 6 EOS 3 19 LBORRES 19 0.25894737
32 ARM D Week 12 EOS 3 20 LBORRES 20 0.25650000
33 ARM A Day 1 HCT 3 20 LBORRES 20 41.84000000
34 ARM A Week 2 HCT 3 20 LBORRES 20 42.18000000
35 ARM A Week 6 HCT 3 19 LBORRES 19 42.46315789
36 ARM A Week 12 HCT 3 19 LBORRES 19 42.26842105
37 ARM B Day 1 HCT 3 21 LBORRES 21 42.33809524
38 ARM B Week 2 HCT 3 20 LBORRES 20 42.86500000
39 ARM B Week 6 HCT 3 18 LBORRES 18 42.23888889
40 ARM B Week 12 HCT 3 18 LBORRES 17 42.48823529
41 ARM C Day 1 HCT 3 19 LBORRES 19 42.48421053
42 ARM C Week 2 HCT 3 18 LBORRES 18 42.50000000
43 ARM C Week 6 HCT 3 17 LBORRES 17 42.28823529
44 ARM C Week 12 HCT 3 16 LBORRES 16 41.83750000
45 ARM D Day 1 HCT 3 23 LBORRES 21 43.05714286
46 ARM D Week 2 HCT 3 22 LBORRES 22 43.42727273
47 ARM D Week 6 HCT 3 20 LBORRES 20 42.91000000
48 ARM D Week 12 HCT 3 20 LBORRES 20 42.69000000
49 ARM A Day 1 HGB 3 20 LBORRES 20 14.45000000
50 ARM A Week 2 HGB 3 20 LBORRES 20 14.52000000
51 ARM A Week 6 HGB 3 19 LBORRES 19 14.65789474
52 ARM A Week 12 HGB 3 19 LBORRES 19 14.51578947
53 ARM B Day 1 HGB 3 21 LBORRES 21 14.60000000
54 ARM B Week 2 HGB 3 20 LBORRES 20 14.74500000
55 ARM B Week 6 HGB 3 18 LBORRES 18 14.60000000
56 ARM B Week 12 HGB 3 18 LBORRES 17 14.71176471
57 ARM C Day 1 HGB 3 19 LBORRES 19 14.54210526
58 ARM C Week 2 HGB 3 18 LBORRES 18 14.58333333
59 ARM C Week 6 HGB 3 17 LBORRES 17 14.50000000
60 ARM C Week 12 HGB 3 16 LBORRES 16 14.36250000
61 ARM D Day 1 HGB 3 23 LBORRES 21 14.83333333
62 ARM D Week 2 HGB 3 22 LBORRES 22 14.91363636
63 ARM D Week 6 HGB 3 20 LBORRES 20 14.83000000
64 ARM D Week 12 HGB 3 20 LBORRES 20 14.75500000
65 ARM A Day 1 LYM 3 19 LBORRES 19 1.94631579
66 ARM A Week 2 LYM 3 19 LBORRES 19 2.00842105
67 ARM A Week 6 LYM 3 19 LBORRES 19 1.90578947
68 ARM A Week 12 LYM 3 18 LBORRES 18 1.93611111
69 ARM B Day 1 LYM 3 20 LBORRES 20 1.84700000
70 ARM B Week 2 LYM 3 20 LBORRES 20 2.00600000
71 ARM B Week 6 LYM 3 18 LBORRES 18 1.82333333
72 ARM B Week 12 LYM 3 18 LBORRES 16 1.89625000
73 ARM C Day 1 LYM 3 19 LBORRES 19 1.77157895
74 ARM C Week 2 LYM 3 18 LBORRES 18 1.88111111
75 ARM C Week 6 LYM 3 17 LBORRES 17 1.80705882
76 ARM C Week 12 LYM 3 15 LBORRES 15 1.78666667
77 ARM D Day 1 LYM 3 25 LBORRES 21 1.94476190
78 ARM D Week 2 LYM 3 22 LBORRES 22 1.84318182
79 ARM D Week 6 LYM 3 19 LBORRES 19 1.85000000
80 ARM D Week 12 LYM 3 20 LBORRES 20 1.77800000
Add mean lab values to change from baseline
Apply formats
=========================================================================
Create report
=========================================================================
Create output path
C:\Users\dbosa\AppData\Local\Temp\RtmpuCtffJ/output/example13.rtf
Create report first so content can be added dynamically
Loop through lab tests
**** Lab Test: Basophils (x10⁹/L) ****
Apply subset
Create plot
Create table for mean change/mean value
Create table for patient counts
Patch together plot and tables
Create plot content
Add content to report
**** Lab Test: Eosinophils (x10⁹/L) ****
Apply subset
Create plot
Create table for mean change/mean value
Create table for patient counts
Patch together plot and tables
Create plot content
Add content to report
**** Lab Test: Hematocrit (%) ****
Apply subset
Create plot
Create table for mean change/mean value
Create table for patient counts
Patch together plot and tables
Create plot content
Add content to report
**** Lab Test: Hemoglobin (g/dL) ****
Apply subset
Create plot
Create table for mean change/mean value
Create table for patient counts
Patch together plot and tables
Create plot content
Add content to report
**** Lab Test: Lymphocytes (x10⁹/L) ****
Apply subset
Create plot
Create table for mean change/mean value
Create table for patient counts
Patch together plot and tables
Create plot content
Add content to report
Write out report to file system
# A report specification: 5 pages
- file_path: 'C:\Users\dbosa\AppData\Local\Temp\RtmpuCtffJ/output/example13.rtf'
- output_type: RTF
- units: inches
- orientation: landscape
- margins: top 0.5 bottom 0.5 left 1 right 1
- line size/count: 9/42
- page_header: left=Sponsor: Archytas right=Study: ABC
- title 1: 'Figure 1.0'
- title 2: 'Mean Change From Baseline (HEMATOLOGY)'
- title 3: 'Safety Population'
- page_footer: left=Date: 02Feb2024 22:44:23 center=Confidential right=Page [pg] of [tpg]
- content:
# A plot specification:
- height: 6
- width: 9
- title 1: 'Laboratory Test: Basophils (x10⁹/L)'
# A plot specification:
- height: 6
- width: 9
- title 1: 'Laboratory Test: Eosinophils (x10⁹/L)'
# A plot specification:
- height: 6
- width: 9
- title 1: 'Laboratory Test: Hematocrit (%)'
# A plot specification:
- height: 6
- width: 9
- title 1: 'Laboratory Test: Hemoglobin (g/dL)'
# A plot specification:
- height: 6
- width: 9
- title 1: 'Laboratory Test: Lymphocytes (x10⁹/L)'
=========================================================================
Clean Up
=========================================================================
=========================================================================
Log End Time: 2024-02-02 22:44:32.173356
Log Elapsed Time: 0 00:00:15
=========================================================================