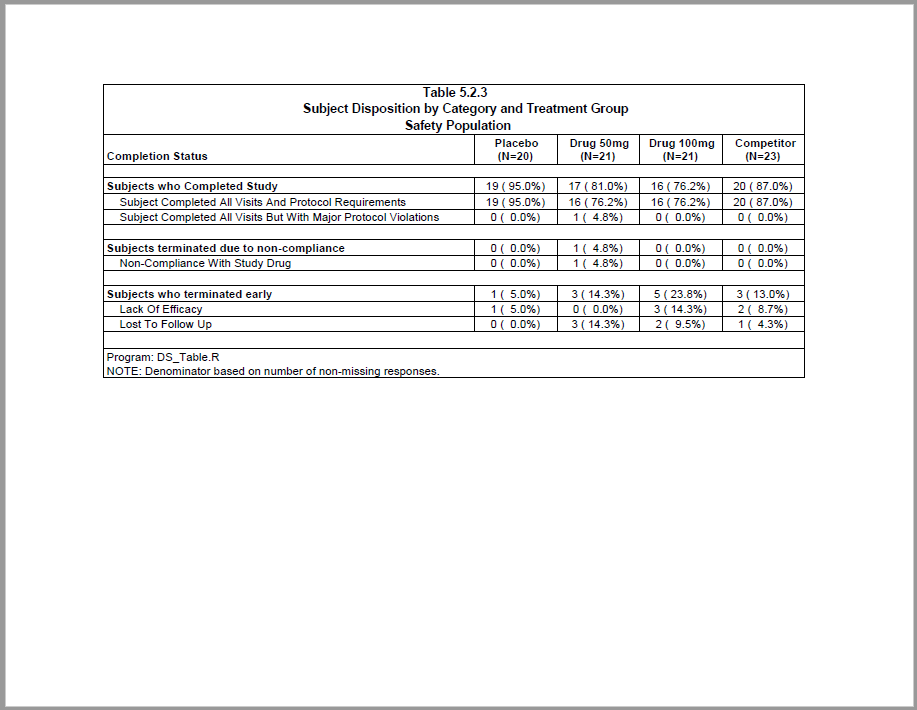
This example produces a subject disposition table. The report features extensive data preparation using the procs package. The example also shows how to create a stub column with summary rows.
Program
library(sassy)
library(stringr)
options("logr.autolog" = TRUE,
"logr.notes" = FALSE,
"logr.on" = TRUE,
"procs.print" = FALSE)
# Get temp directory
tmp <- tempdir()
# Open log
lf <- log_open(file.path(tmp, "example10.log"))
# Get data
dir <- system.file("extdata", package = "sassy")
# Load and Prepare Data ---------------------------------------------------
sep("Prepare Data")
put("Define data library")
libname(sdtm, dir, "csv")
put("Prepare DM data")
datastep(sdtm$DM, keep = v(USUBJID, ARM),
where = expression(ARM != "SCREEN FAILURE"), {}) -> dm_mod
put("Prepare DS data")
datastep(sdtm$DS, keep = v(USUBJID, DSTERM, DSDECOD, DSCAT),
where = expression(DSCAT != "PROTOCOL MILESTONE"), {}) -> ds_mod
put("Join DM with DS to get ARMs on DS")
datastep(dm_mod, merge = ds_mod, merge_by = USUBJID, {}) -> dmds
put("Change ARM to factor to assist with sparse data")
dmds$ARM <- factor(dmds$ARM, levels = c("ARM A", "ARM B", "ARM C", "ARM D"))
put("Get ARM population counts")
proc_freq(dm_mod, tables = ARM, output = long,
options = v(nonobs, nopercent)) -> arm_pop
# Prepare formats ---------------------------------------------------------
# Completed Study
complete_fmt <- value(condition(x == "SUBJECT COMPLETED ALL VISITS AND PROTOCOL REQUIREMENTS",
str_to_title("SUBJECT COMPLETED ALL VISITS AND PROTOCOL REQUIREMENTS")),
condition(x == "SUBJECT COMPLETED ALL VISITS BUT WITH MAJOR PROTOCOL VIOLATIONS",
str_to_title("SUBJECT COMPLETED ALL VISITS BUT WITH MAJOR PROTOCOL VIOLATIONS")))
# Subject Non-compliance
noncomp_fmt <- value(condition(x == "NON-COMPLIANCE WITH STUDY DRUG",
str_to_title("NON-COMPLIANCE WITH STUDY DRUG")))
# Early Termination
term_fmt <- value(condition(x == "LACK OF EFFICACY",
str_to_title("LACK OF EFFICACY")),
condition(str_detect(x, "LOST"),
str_to_title("LOST TO FOLLOW UP")),
condition(TRUE, str_to_title("LACK OF EFFICACY")))
# Group labels
group_fmt <- value(condition(x == "COMPLETED", "Subjects who Completed Study"),
condition(x == "NONCOMPLIANCE", "Subjects terminated due to non-compliance"),
condition(x == "OTHER", "Subjects who terminated early"))
# Disposition Groups ------------------------------------------------------
put("Create vector of final dataframe columns")
cols <- v(group, cat, catseq, `ARM A`, `ARM B`, `ARM C`, `ARM D`) |> put()
put("Get group counts")
proc_freq(dmds, tables = DSDECOD, by = ARM) |>
datastep(keep = v(BY, CAT, CNTPCT), {
CNTPCT <- fmt_cnt_pct(CNT, arm_pop[[BY]])
}) |>
proc_transpose(var = CNTPCT, by = CAT, id = BY) |>
datastep(keep = cols,
{
group <- ifelse(CAT == "NON-COMPLIANCE WITH STUDY DRUG", "NONCOMPLIANCE", CAT)
cat = NA
catseq = 1
}) -> grps
# Disposition Subgroups ----------------------------------------------------
put("Pull out subjects who completed study.")
datastep(dmds, where = expression(DSDECOD == "COMPLETED"),
{
TERMDECOD <- fapply(DSTERM, complete_fmt)
}) |>
proc_freq(tables = v(DSDECOD * TERMDECOD), by = ARM) |>
datastep(keep = v(BY, CAT1, CAT2, CNTPCT),
{
CNTPCT <- fmt_cnt_pct(CNT, arm_pop[[BY]])
}) |>
proc_transpose(var = CNTPCT, by = v(CAT1, CAT2), id = BY) |>
datastep(keep = cols,
{
group = CAT1
cat = CAT2
catseq = 2
}) -> cmplt
put("Pull out subjects who were non-compliant")
datastep(dmds, where = expression(DSDECOD == "NON-COMPLIANCE WITH STUDY DRUG"),
{
TERMDECOD <- fapply(DSTERM, noncomp_fmt)
}) |>
proc_freq(tables = v(DSDECOD * TERMDECOD), by = ARM) |>
datastep(keep = v(BY, CAT1, CAT2, CNTPCT),
{
CNTPCT <- fmt_cnt_pct(CNT, arm_pop[[BY]])
}) |>
proc_transpose(var = CNTPCT, by = v(CAT1, CAT2), id = BY) |>
datastep(keep = cols,
{
group = "NONCOMPLIANCE"
cat = CAT2
catseq = 2
}) -> noncompl
put("Pull out subjects who terminated early")
datastep(dmds, where = expression(DSDECOD == "OTHER"),
{
TERMDECOD <- fapply(DSTERM, term_fmt)
}) |>
proc_freq(tables = v(DSDECOD * TERMDECOD), by = ARM) |>
datastep(keep = v(BY, CAT1, CAT2, CNTPCT),
{
CNTPCT <- fmt_cnt_pct(CNT, arm_pop[[BY]])
}) |>
proc_transpose(var = CNTPCT, by = v(CAT1, CAT2), id = BY) |>
datastep(keep = cols,
{
group = "OTHER"
cat = CAT2
catseq = 2
}) -> earlyterm
put("Combine blocks into final data frame")
datastep(grps, set = list(cmplt, noncompl, earlyterm),
{
lblind <- ifelse(is.na(cat), TRUE, FALSE)
}) |>
proc_sort(by = v(group, catseq, cat)) -> final
# Report ------------------------------------------------------------------
sep("Create and print report")
# Create Table
tbl <- create_table(final, first_row_blank = TRUE,
borders = "all", width = 8.5, header_bold = TRUE) |>
column_defaults(from = `ARM A`, to = `ARM D`,
align = "center", width = 1) |>
stub(vars = v(group, cat), "Completion Status",
style = cell_style(bold = TRUE, indicator = "lblind")) |>
define(group, blank_after = TRUE, dedupe = TRUE,
format = group_fmt) |>
define(cat, indent = .5) |>
define(catseq, visible = FALSE) |>
define(`ARM A`, label = "Placebo", n = arm_pop["ARM A"]) |>
define(`ARM B`, label = "Drug 50mg", n = arm_pop["ARM B"]) |>
define(`ARM C`, label = "Drug 100mg", n = arm_pop["ARM C"]) |>
define(`ARM D`, label = "Competitor", n = arm_pop["ARM D"]) |>
define(lblind, visible = FALSE) |>
titles("Table 5.2.3", "Subject Disposition by Category and Treatment Group",
"Safety Population", bold = TRUE, font_size = 11,
borders = "outside", blank_row = "none") |>
footnotes("Program: DS_Table.R",
"NOTE: Denominator based on number of non-missing responses.",
borders = "outside", blank_row = "none")
pth <- file.path(tmp, "example10.pdf")
rpt <- create_report(pth, output_type = "PDF", font = "Arial") |>
set_margins(top = 1, bottom = 1) |>
add_content(tbl)
write_report(rpt)
# Clean Up ----------------------------------------------------------------
# Close log
log_close()
# Uncomment to view files
# file.show(pth)
# file.show(lf)Log
Here is the log:
=========================================================================
Log Path: C:/Users/dbosa/AppData/Local/Temp/RtmpEBPgPu/log/example10.log
Program Path: C:/packages/Testing/procs/ProcsDisposition.R
Working Directory: C:/packages/Testing/procs
User Name: dbosa
R Version: 4.3.1 (2023-06-16 ucrt)
Machine: SOCRATES x86-64
Operating System: Windows 10 x64 build 22621
Base Packages: stats graphics grDevices utils datasets methods base Other
Packages: tidylog_1.0.2 stringr_1.5.0 procs_1.0.3 reporter_1.4.1 libr_1.2.8
fmtr_1.5.9 logr_1.3.4 common_1.0.8 sassy_1.1.0
Log Start Time: 2023-09-05 22:06:05.090672
=========================================================================
=========================================================================
Prepare Data
=========================================================================
Define data library
# library 'sdtm': 7 items
- attributes: csv not loaded
- path: C:/Users/dbosa/AppData/Local/R/win-library/4.3/sassy/extdata
- items:
Name Extension Rows Cols Size
1 AE csv 150 27 88.5 Kb
2 DM csv 87 24 45.5 Kb
3 DS csv 174 9 34.1 Kb
4 EX csv 84 11 26.4 Kb
5 IE csv 2 14 13.4 Kb
6 SV csv 685 10 70.3 Kb
7 VS csv 3358 17 467.4 Kb
LastModified
1 2023-08-07 17:51:40
2 2023-08-07 17:51:40
3 2023-08-07 17:51:40
4 2023-08-07 17:51:40
5 2023-08-07 17:51:40
6 2023-08-07 17:51:40
7 2023-08-07 17:51:40
Prepare DM data
datastep: columns decreased from 24 to 2
# A tibble: 85 × 2
USUBJID ARM
<chr> <chr>
1 ABC-01-049 ARM D
2 ABC-01-050 ARM B
3 ABC-01-051 ARM A
4 ABC-01-052 ARM C
5 ABC-01-053 ARM B
6 ABC-01-054 ARM D
7 ABC-01-055 ARM C
8 ABC-01-056 ARM A
9 ABC-01-113 ARM D
10 ABC-01-114 ARM B
# ℹ 75 more rows
# ℹ Use `print(n = ...)` to see more rows
Prepare DS data
datastep: columns decreased from 9 to 4
# A tibble: 87 × 4
USUBJID DSTERM DSDECOD DSCAT
<chr> <chr> <chr> <chr>
1 ABC-01-049 SUBJECT COMPLETED ALL … COMPLE… DISP…
2 ABC-01-050 SUBJECT COMPLETED ALL … COMPLE… DISP…
3 ABC-01-051 SUBJECT COMPLETED ALL … COMPLE… DISP…
4 ABC-01-052 SUBJECT COMPLETED ALL … COMPLE… DISP…
5 ABC-01-053 NON-COMPLIANCE WITH ST… NON-CO… DISP…
6 ABC-01-054 SUBJECT COMPLETED ALL … COMPLE… DISP…
7 ABC-01-055 SUBJECT COMPLETED ALL … COMPLE… DISP…
8 ABC-01-056 SUBJECT COMPLETED ALL … COMPLE… DISP…
9 ABC-01-113 SUBJECT COMPLETED ALL … COMPLE… DISP…
10 ABC-01-114 SUBJECT COMPLETED ALL … COMPLE… DISP…
# ℹ 77 more rows
# ℹ Use `print(n = ...)` to see more rows
Join DM with DS to get ARMs on DS
datastep: columns increased from 2 to 5
# A tibble: 87 × 5
USUBJID ARM DSTERM DSDECOD DSCAT
<chr> <chr> <chr> <chr> <chr>
1 ABC-01-049 ARM D SUBJECT COMPLETE… COMPLE… DISP…
2 ABC-01-050 ARM B SUBJECT COMPLETE… COMPLE… DISP…
3 ABC-01-051 ARM A SUBJECT COMPLETE… COMPLE… DISP…
4 ABC-01-052 ARM C SUBJECT COMPLETE… COMPLE… DISP…
5 ABC-01-053 ARM B NON-COMPLIANCE W… NON-CO… DISP…
6 ABC-01-054 ARM D SUBJECT COMPLETE… COMPLE… DISP…
7 ABC-01-055 ARM C SUBJECT COMPLETE… COMPLE… DISP…
8 ABC-01-056 ARM A SUBJECT COMPLETE… COMPLE… DISP…
9 ABC-01-113 ARM D SUBJECT COMPLETE… COMPLE… DISP…
10 ABC-01-114 ARM B SUBJECT COMPLETE… COMPLE… DISP…
# ℹ 77 more rows
# ℹ Use `print(n = ...)` to see more rows
Change ARM to factor to assist with sparse data
Get ARM population counts
proc_freq: input data set 85 rows and 2 columns
tables: ARM
output: long
view: TRUE
output: 1 datasets
# A tibble: 1 × 6
VAR STAT `ARM A` `ARM B` `ARM C` `ARM D`
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 ARM CNT 20 21 21 23
# A user-defined format: 2 conditions
Name Type
1 obj U
2 obj U
Expression
1 x == "SUBJECT COMPLETED ALL VISITS AND PROTOCOL REQUIREMENTS"
2 x == "SUBJECT COMPLETED ALL VISITS BUT WITH MAJOR PROTOCOL VIOLATIONS"
Label
1 Subject Completed All Visits And Protocol Requirements
2 Subject Completed All Visits But With Major Protocol Violations
Order
1 NA
2 NA
# A user-defined format: 1 conditions
Name Type Expression
1 obj U x == "NON-COMPLIANCE WITH STUDY DRUG"
Label Order
1 Non-Compliance With Study Drug NA
# A user-defined format: 3 conditions
Name Type Expression
1 obj U x == "LACK OF EFFICACY"
2 obj U str_detect(x, "LOST")
3 obj U TRUE
Label Order
1 Lack Of Efficacy NA
2 Lost To Follow Up NA
3 Lack Of Efficacy NA
# A user-defined format: 3 conditions
Name Type Expression
1 obj U x == "COMPLETED"
2 obj U x == "NONCOMPLIANCE"
3 obj U x == "OTHER"
Label Order
1 Subjects who Completed Study NA
2 Subjects terminated due to non-compliance NA
3 Subjects who terminated early NA
Create vector of final dataframe columns
group
cat
catseq
ARM A
ARM B
ARM C
ARM D
Get group counts
proc_freq: input data set 87 rows and 5 columns
tables: DSDECOD
by: ARM
view: TRUE
output: 1 datasets
# A tibble: 12 × 6
BY VAR CAT N CNT PCT
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 ARM A DSDECOD COMPLETED 20 19 95
2 ARM A DSDECOD NON-COMPLIANCE … 20 0 0
3 ARM A DSDECOD OTHER 20 1 5
4 ARM B DSDECOD COMPLETED 21 17 81.0
5 ARM B DSDECOD NON-COMPLIANCE … 21 1 4.76
6 ARM B DSDECOD OTHER 21 3 14.3
7 ARM C DSDECOD COMPLETED 21 16 76.2
8 ARM C DSDECOD NON-COMPLIANCE … 21 0 0
9 ARM C DSDECOD OTHER 21 5 23.8
10 ARM D DSDECOD COMPLETED 23 20 87.0
11 ARM D DSDECOD NON-COMPLIANCE … 23 0 0
12 ARM D DSDECOD OTHER 23 3 13.0
datastep: columns decreased from 6 to 3
# A tibble: 12 × 3
BY CAT CNTPCT
<chr> <chr> <chr>
1 ARM A COMPLETED 19 ( 95.0%)
2 ARM A NON-COMPLIANCE WITH STUDY DRUG 0 ( 0.0%)
3 ARM A OTHER 1 ( 5.0%)
4 ARM B COMPLETED 17 ( 81.0%)
5 ARM B NON-COMPLIANCE WITH STUDY DRUG 1 ( 4.8%)
6 ARM B OTHER 3 ( 14.3%)
7 ARM C COMPLETED 16 ( 76.2%)
8 ARM C NON-COMPLIANCE WITH STUDY DRUG 0 ( 0.0%)
9 ARM C OTHER 5 ( 23.8%)
10 ARM D COMPLETED 20 ( 87.0%)
11 ARM D NON-COMPLIANCE WITH STUDY DRUG 0 ( 0.0%)
12 ARM D OTHER 3 ( 13.0%)
proc_transpose: input data set 12 rows and 3 columns
by: CAT
var: CNTPCT
id: BY
name: NAME
output dataset 3 rows and 6 columns
# A tibble: 3 × 6
CAT NAME `ARM A` `ARM B` `ARM C` `ARM D`
<chr> <chr> <chr> <chr> <chr> <chr>
1 COMPLETED CNTP… 19 ( 9… 17 ( 8… 16 ( 7… 20 ( 8…
2 NON-COMPLI… CNTP… 0 ( 0… 1 ( 4… 0 ( 0… 0 ( 0…
3 OTHER CNTP… 1 ( 5… 3 ( 14… 5 ( 23… 3 ( 13…
datastep: columns increased from 6 to 7
# A tibble: 3 × 7
group cat catseq `ARM A` `ARM B` `ARM C`
<chr> <lgl> <dbl> <chr> <chr> <chr>
1 COMPLETED NA 1 19 ( 9… 17 ( 8… 16 ( 7…
2 NONCOMPLIAN… NA 1 0 ( 0… 1 ( 4… 0 ( 0…
3 OTHER NA 1 1 ( 5… 3 ( 14… 5 ( 23…
# ℹ 1 more variable: `ARM D` <chr>
Pull out subjects who completed study.
datastep: columns increased from 5 to 6
# A tibble: 74 × 6
USUBJID ARM DSTERM DSDECOD DSCAT TERMDECOD
<chr> <fct> <chr> <chr> <chr> <chr>
1 ABC-01-049 ARM D SUBJEC… COMPLE… DISP… Subject …
2 ABC-01-050 ARM B SUBJEC… COMPLE… DISP… Subject …
3 ABC-01-051 ARM A SUBJEC… COMPLE… DISP… Subject …
4 ABC-01-052 ARM C SUBJEC… COMPLE… DISP… Subject …
5 ABC-01-054 ARM D SUBJEC… COMPLE… DISP… Subject …
6 ABC-01-055 ARM C SUBJEC… COMPLE… DISP… Subject …
7 ABC-01-056 ARM A SUBJEC… COMPLE… DISP… Subject …
8 ABC-01-113 ARM D SUBJEC… COMPLE… DISP… Subject …
9 ABC-01-114 ARM B SUBJEC… COMPLE… DISP… Subject …
10 ABC-02-033 ARM C SUBJEC… COMPLE… DISP… Subject …
# ℹ 64 more rows
# ℹ Use `print(n = ...)` to see more rows
proc_freq: input data set 74 rows and 6 columns
tables: DSDECOD * TERMDECOD
by: ARM
view: TRUE
output: 1 datasets
# A tibble: 8 × 8
BY VAR1 VAR2 CAT1 CAT2 N CNT PCT
<chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 ARM A DSDEC… TERM… COMP… Subj… 19 19 100
2 ARM A DSDEC… TERM… COMP… Subj… 19 0 0
3 ARM B DSDEC… TERM… COMP… Subj… 17 16 94.1
4 ARM B DSDEC… TERM… COMP… Subj… 17 1 5.88
5 ARM C DSDEC… TERM… COMP… Subj… 16 16 100
6 ARM C DSDEC… TERM… COMP… Subj… 16 0 0
7 ARM D DSDEC… TERM… COMP… Subj… 20 20 100
8 ARM D DSDEC… TERM… COMP… Subj… 20 0 0
datastep: columns decreased from 8 to 4
# A tibble: 8 × 4
BY CAT1 CAT2 CNTPCT
<chr> <chr> <chr> <chr>
1 ARM A COMPLETED Subject Completed All Vis… 19 ( …
2 ARM A COMPLETED Subject Completed All Vis… 0 ( …
3 ARM B COMPLETED Subject Completed All Vis… 16 ( …
4 ARM B COMPLETED Subject Completed All Vis… 1 ( …
5 ARM C COMPLETED Subject Completed All Vis… 16 ( …
6 ARM C COMPLETED Subject Completed All Vis… 0 ( …
7 ARM D COMPLETED Subject Completed All Vis… 20 ( …
8 ARM D COMPLETED Subject Completed All Vis… 0 ( …
proc_transpose: input data set 8 rows and 4 columns
by: CAT1 CAT2
var: CNTPCT
id: BY
name: NAME
output dataset 2 rows and 7 columns
# A tibble: 2 × 7
CAT1 CAT2 NAME `ARM A` `ARM B` `ARM C` `ARM D`
<chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 COMP… Subj… CNTP… 19 ( 9… 16 ( 7… 16 ( 7… 20 ( 8…
2 COMP… Subj… CNTP… 0 ( 0… 1 ( 4… 0 ( 0… 0 ( 0…
datastep: columns started with 7 and ended with 7
# A tibble: 2 × 7
group cat catseq `ARM A` `ARM B` `ARM C`
<chr> <chr> <dbl> <chr> <chr> <chr>
1 COMPLETED Subject… 2 19 ( 9… 16 ( 7… 16 ( 7…
2 COMPLETED Subject… 2 0 ( 0… 1 ( 4… 0 ( 0…
# ℹ 1 more variable: `ARM D` <chr>
Pull out subjects who were non-compliant
datastep: columns increased from 5 to 6
# A tibble: 1 × 6
USUBJID ARM DSTERM DSDECOD DSCAT TERMDECOD
<chr> <fct> <chr> <chr> <chr> <chr>
1 ABC-01-053 ARM B NON-COM… NON-CO… DISP… Non-Comp…
proc_freq: input data set 1 rows and 6 columns
tables: DSDECOD * TERMDECOD
by: ARM
view: TRUE
output: 1 datasets
# A tibble: 4 × 8
BY VAR1 VAR2 CAT1 CAT2 N CNT PCT
<chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 ARM A DSDECOD TERM… NON-… Non-… 0 0 NaN
2 ARM B DSDECOD TERM… NON-… Non-… 1 1 100
3 ARM C DSDECOD TERM… NON-… Non-… 0 0 NaN
4 ARM D DSDECOD TERM… NON-… Non-… 0 0 NaN
datastep: columns decreased from 8 to 4
# A tibble: 4 × 4
BY CAT1 CAT2 CNTPCT
<chr> <chr> <chr> <chr>
1 ARM A NON-COMPLIANCE WITH STUDY DRUG Non-… 0 ( …
2 ARM B NON-COMPLIANCE WITH STUDY DRUG Non-… 1 ( …
3 ARM C NON-COMPLIANCE WITH STUDY DRUG Non-… 0 ( …
4 ARM D NON-COMPLIANCE WITH STUDY DRUG Non-… 0 ( …
proc_transpose: input data set 4 rows and 4 columns
by: CAT1 CAT2
var: CNTPCT
id: BY
name: NAME
output dataset 1 rows and 7 columns
# A tibble: 1 × 7
CAT1 CAT2 NAME `ARM A` `ARM B` `ARM C` `ARM D`
<chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 NON-… Non-… CNTP… 0 ( 0… 1 ( 4… 0 ( 0… 0 ( 0…
datastep: columns started with 7 and ended with 7
# A tibble: 1 × 7
group cat catseq `ARM A` `ARM B` `ARM C`
<chr> <chr> <dbl> <chr> <chr> <chr>
1 NONCOMPLIAN… Non-… 2 0 ( 0… 1 ( 4… 0 ( 0…
# ℹ 1 more variable: `ARM D` <chr>
Pull out subjects who terminated early
datastep: columns increased from 5 to 6
# A tibble: 12 × 6
USUBJID ARM DSTERM DSDECOD DSCAT TERMDECOD
<chr> <fct> <chr> <chr> <chr> <chr>
1 ABC-03-005 ARM C LOST O… OTHER DISP… Lost To …
2 ABC-03-008 ARM D WORSEN… OTHER DISP… Lack Of …
3 ABC-04-074 ARM C LACK O… OTHER DISP… Lack Of …
4 ABC-04-128 ARM C LOST T… OTHER DISP… Lost To …
5 ABC-06-069 ARM A PSORIA… OTHER DISP… Lack Of …
6 ABC-06-161 ARM C NON R… OTHER DISP… Lack Of …
7 ABC-08-103 ARM B LOST T… OTHER DISP… Lost To …
8 ABC-08-105 ARM C MOTHER… OTHER DISP… Lack Of …
9 ABC-08-107 ARM D PATIEN… OTHER DISP… Lack Of …
10 ABC-08-108 ARM B LOST T… OTHER DISP… Lost To …
11 ABC-09-021 ARM B LOST T… OTHER DISP… Lost To …
12 ABC-09-022 ARM D LOST T… OTHER DISP… Lost To …
proc_freq: input data set 12 rows and 6 columns
tables: DSDECOD * TERMDECOD
by: ARM
view: TRUE
output: 1 datasets
# A tibble: 8 × 8
BY VAR1 VAR2 CAT1 CAT2 N CNT PCT
<chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 ARM A DSDECOD TERM… OTHER Lack… 1 1 100
2 ARM A DSDECOD TERM… OTHER Lost… 1 0 0
3 ARM B DSDECOD TERM… OTHER Lack… 3 0 0
4 ARM B DSDECOD TERM… OTHER Lost… 3 3 100
5 ARM C DSDECOD TERM… OTHER Lack… 5 3 60
6 ARM C DSDECOD TERM… OTHER Lost… 5 2 40
7 ARM D DSDECOD TERM… OTHER Lack… 3 2 66.7
8 ARM D DSDECOD TERM… OTHER Lost… 3 1 33.3
datastep: columns decreased from 8 to 4
# A tibble: 8 × 4
BY CAT1 CAT2 CNTPCT
<chr> <chr> <chr> <chr>
1 ARM A OTHER Lack Of Efficacy 1 ( 5.0%)
2 ARM A OTHER Lost To Follow Up 0 ( 0.0%)
3 ARM B OTHER Lack Of Efficacy 0 ( 0.0%)
4 ARM B OTHER Lost To Follow Up 3 ( 14.3%)
5 ARM C OTHER Lack Of Efficacy 3 ( 14.3%)
6 ARM C OTHER Lost To Follow Up 2 ( 9.5%)
7 ARM D OTHER Lack Of Efficacy 2 ( 8.7%)
8 ARM D OTHER Lost To Follow Up 1 ( 4.3%)
proc_transpose: input data set 8 rows and 4 columns
by: CAT1 CAT2
var: CNTPCT
id: BY
name: NAME
output dataset 2 rows and 7 columns
# A tibble: 2 × 7
CAT1 CAT2 NAME `ARM A` `ARM B` `ARM C` `ARM D`
<chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 OTHER Lack… CNTP… 1 ( 5… 0 ( 0… 3 ( 14… 2 ( 8…
2 OTHER Lost… CNTP… 0 ( 0… 3 ( 14… 2 ( 9… 1 ( 4…
datastep: columns started with 7 and ended with 7
# A tibble: 2 × 7
group cat catseq `ARM A` `ARM B` `ARM C`
<chr> <chr> <dbl> <chr> <chr> <chr>
1 OTHER Lack Of Eff… 2 1 ( 5… 0 ( 0… 3 ( 14…
2 OTHER Lost To Fol… 2 0 ( 0… 3 ( 14… 2 ( 9…
# ℹ 1 more variable: `ARM D` <chr>
Combine blocks into final data frame
datastep: columns increased from 7 to 8
# A tibble: 8 × 8
group cat catseq `ARM A` `ARM B` `ARM C`
<chr> <chr> <dbl> <chr> <chr> <chr>
1 COMPLETED <NA> 1 19 ( 9… 17 ( 8… 16 ( 7…
2 NONCOMPLIAN… <NA> 1 0 ( 0… 1 ( 4… 0 ( 0…
3 OTHER <NA> 1 1 ( 5… 3 ( 14… 5 ( 23…
4 COMPLETED Subj… 2 19 ( 9… 16 ( 7… 16 ( 7…
5 COMPLETED Subj… 2 0 ( 0… 1 ( 4… 0 ( 0…
6 NONCOMPLIAN… Non-… 2 0 ( 0… 1 ( 4… 0 ( 0…
7 OTHER Lack… 2 1 ( 5… 0 ( 0… 3 ( 14…
8 OTHER Lost… 2 0 ( 0… 3 ( 14… 2 ( 9…
# ℹ 2 more variables: `ARM D` <chr>, lblind <lgl>
proc_sort: input data set 8 rows and 8 columns
by: group catseq cat
keep: group cat catseq ARM A ARM B ARM C ARM D lblind
order: a a a
output data set 8 rows and 8 columns
# A tibble: 8 × 8
group cat catseq `ARM A` `ARM B` `ARM C`
<chr> <chr> <dbl> <chr> <chr> <chr>
1 COMPLETED <NA> 1 19 ( 9… 17 ( 8… 16 ( 7…
2 COMPLETED Subj… 2 19 ( 9… 16 ( 7… 16 ( 7…
3 COMPLETED Subj… 2 0 ( 0… 1 ( 4… 0 ( 0…
4 NONCOMPLIAN… <NA> 1 0 ( 0… 1 ( 4… 0 ( 0…
5 NONCOMPLIAN… Non-… 2 0 ( 0… 1 ( 4… 0 ( 0…
6 OTHER <NA> 1 1 ( 5… 3 ( 14… 5 ( 23…
7 OTHER Lack… 2 1 ( 5… 0 ( 0… 3 ( 14…
8 OTHER Lost… 2 0 ( 0… 3 ( 14… 2 ( 9…
# ℹ 2 more variables: `ARM D` <chr>, lblind <lgl>
=========================================================================
Create and print report
=========================================================================
# A report specification: 1 pages
- file_path: 'C:\Users\dbosa\AppData\Local\Temp\RtmpEBPgPu/example10.pdf'
- output_type: PDF
- units: inches
- orientation: landscape
- margins: top 1 bottom 1 left 1 right 1
- line size/count: 9/41
- content:
# A table specification:
- data: tibble 'final' 8 rows 8 cols
- show_cols: all
- use_attributes: all
- width: 8.5
- title 1: 'Table 5.2.3'
- title 2: 'Subject Disposition by Category and Treatment Group'
- title 3: 'Safety Population'
- footnote 1: 'Program: DS_Table.R'
- footnote 2: 'NOTE: Denominator based on number of non-missing responses.'
- stub: group cat 'Completion Status' align='left'
- define: group dedupe='TRUE'
- define: cat
- define: catseq visible='FALSE'
- define: ARM A 'Placebo'
- define: ARM B 'Drug 50mg'
- define: ARM C 'Drug 100mg'
- define: ARM D 'Competitor'
- define: lblind visible='FALSE'
lib_sync: synchronized data in library 'sdtm'
=========================================================================
Log End Time: 2023-09-05 22:06:09.329367
Log Elapsed Time: 0 00:00:04
=========================================================================